DIFSIM
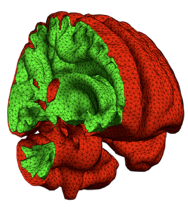
DIFSIM is CSCI’s numerical simulation software for diffusion MRI data. It can accurately model the MR diffusion signal within any given tissues geometry and provides a simulation environment for modeling and testing an entire diffusion MRI experiment. Individual molecules (spins) are tracked as they diffuse through the arbitrarily complex 3D environment and through applied magnetic fields. Molecular diffusion is simulated using the MCell simulator of microcellular physiology. As a result, DIFSIM facilitates a practical approach to carrying out virtual experiments to explore the full parameter space, which can be adapted to validate, standardize, and optimize DTI acquisition methods for quantitative consistency. Recent work has involved the development of a whole brain simulation with computational grids derived from actual high resolution dFPG anatomical MRI data. This is now being used to develop gray matter diffusion MRI for use in the detection of traumatic brain injury (TBI).
DIFSIM was also recently used to establish a series of relationships that defined how muscle microstructure and DTI are related. This novel in silico method identified features of muscle microstructure that DTI is most sensitive to, and which combinations are beyond the sensitivity of the technique, providing the necessary experimental control to uncouple muscle microstructure from diffusion imaging.
If you would like to request a copy of the DIFSIM software for testing and development, please contact us via email for download instructions at:
