Morphology
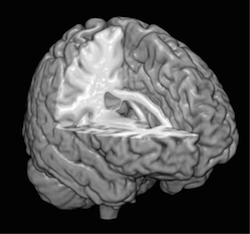
We utilize a novel approach based on spherical wave decomposition (SWD) [1] that we developed to overcome the limitations of surface-based methods by directly analyzing the entire data volume, obviating the segmentation, inflation, and surface fitting steps, significantly reducing the computational time and eliminating topological errors while providing a more detailed quantitative description based upon a more complete theoretical framework for volumetric data. The SWD approach provides a precise quantitative description of the morphology that encapsulates the geometric complexity of the brain and facilitates the determination of important quantitative metrics of brain structure, such as a global measure of cortical foliation, derived from the optimal order of SWD fit, to the local measurements of cortical thickness, which provides critical clinically relevant information in a wide range of brain diseases.
Diffusion Tensor Imaging analysis
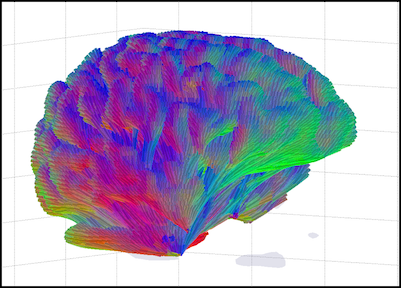
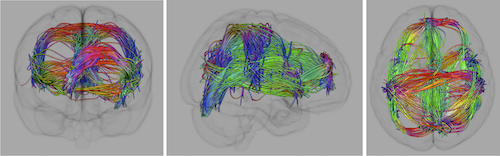
We have developed a novel approach to the analysis of DTI data in which we use local coupling information, in conjunction with a geometric optics based tract tracing method, to simultaneous estimate both the local diffusion and global fiber tracts [2]. This method uses our recently develop theory of Entropy Spectrum Pathways (ESP) [3] theory to incorporate local coupling between sub–scale diffusion parameters in order to compute the structure of the equilibrium probabilities that define the global information entropy field and uses this global entropy to update the local properties of neural fiber tracts. This approach has an advantage over existing methods in that it does not depend on any model for the number of fibers and can easily distinguish many (e.g., 7) fibers traversing a voxel. In addition, we have also developed an efficient way to trace individual tracts that utilizes the multi–scale and multi–modal structure by means of an approach reminiscent of the geometrical optics ray tracing in dispersive media (either elastic or viscoelastic). This geometrical optics–like approach naturally includes multiple scales that allows fiber tracing to continue through voxels with complex local diffusion properties where multiple fiber directions are unable to be adequately resolved. We call this method GO-ESP [2].
Resting state FMRI analysis
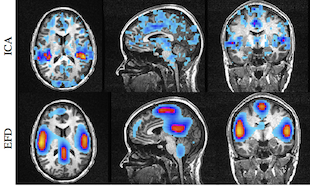
Our approach to resting state FMRI analysis is our recently developed entropy field decomposition (EFD) method [4][5] that develops a field-theoretic description of probability theory, supplemented by prior information provided by our ESP formulation [2]. This method is based upon a general information field theoretic formulation of Bayesian probability theory incorporating prior coupling information that allows the enumeration of the most probable parameter configurations without the need for unjustified statistical assumptions. The result is a method by which space-time correlations in the data can be used to detect modes of activity and rank them according to their significance (e.g., their power). The non-Gaussian and non-linear features of this method are important for rsFMRI data, which is expected to have these characteristics as well. This approach facilitates the construction of brain activation modes directly from the spatial-temporal correlation structure of the data. Importantly, the EFD procedure is not limited to independent modes, but can characterize interactions between space-time modes as well, a feature we believe to be important in the characterization of brain activity.
We have also developed a novel method for tracking these modes and their associated spatial-temporal correlation structure can then be used to generate space-time activity probability trajectories called functional connectivity pathways which provide a characterization of functional brain networks.
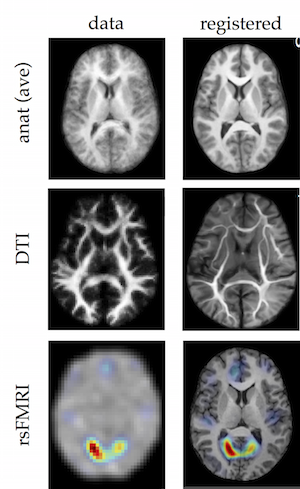
Image registration
We have developed a novel Hamiltonian formalism to non-linear flexible image registration. The method builds a diffeomorphic map as a sequence of symplectomorphic maps with each map embedded in a separate energy shell. The approach adds a novel phase space regularization based on a powerful entropy spectrum pathways framework. The framework provides a unique opportunity to tailor image details into a regularization scheme by choosing an image derived regularization kernel. Spherical wave decomposition is applied as a powerful preconditioning tool in a position domain allowing accurate and fast interpolation, resampling and estimation of fixed shape rotation and scale. The result is an efficient and versatile method capable of fast and accurate registration of a variety of volumetric images of different modalities and resolutions.
References:
- V. L. Galinsky and L. R. Frank, “Automated segmentation and shape characterization of volumetric data,” NeuroImage, vol. 92, pp. 156–168, May 2014.
- L. R. Frank and V. L. Galinsky, “Information pathways in a disordered lattice,” Physical Review E, vol. 89, p. 032142, Mar 2014.
- V. L. Galinsky and L. R. Frank, “Simultaneous multi-scale diffusion estimation and tractography guided by entropy spectrum pathways,” IEEE Transactions on Medical Imaging, vol. PP, no. 99, pp. 1–1, 2014.
- L. R. Frank and V. L. Galinsky, “Dynamic multi-scale modes of resting state brain activity detected by entropy field decomposition,” Neural Computation, 28(9): 1769-1811, 2016.
- L. R. Frank and V. L. Galinsky. "Detecting spatio-temporal modes in multivariate data by entropy field decomposition," Journal of Physics A, 49: 395001, 2016.